Institute of Advanced Materials (INAM)
Universitat Jaume I
12006 Castelló
Spain
Publications
2025
-
Nature Methods, 2025, 22, 641.
Amaro, R.E.; Åqvist, J.; Bahar, I.; Battistini, F.; Bellaiche, A.; Beltran, D.; Biggin, P.C.; Bonomi, M.; Bowman, G.R.; Bryce, R.; Bussi, G.; Carloni, P.; Case, D.; Cavalli, A.; Chang, C.E.A.; Cheung, M.S.; Chipot, C.; Chong, L.T.; Choudhary, P.; Cisneros, G.A.; Clementi, C.; Collepardo-Guevara, R.; Coveney, P.; Covino, R.; Crawford, T.D.; Peraro, M.D.; de Groot, B.; Delemotte, L.; Vivo, M.D.; Essex, J.; Fraternali, F.; Gao, J.; Gelpí, J.L.; Gervasio, F.L.; Gonzalez-Nilo, F.D.; Grubmüller, H.; Guenza, M.; Guzman, H.V.; Harris, S.; Head-Gordon, T.; Hernandez, R.; Hospital, A.; Huang, N.; Huang, X.; Hummer, G.; Iglesias-Fernández, J.; Jensen, J.H.; Jha, S.; Jiao, W.; Jorgensen, W.L.; Kamerlin, S.C.L.; Khalid, S.; Laughton, C.; Levitt, M.; Limongelli, V.; Lindahl, E.; Lindorff-Larsen, K.; Loverde, S.; Lundborg, M.; Luo, Y.L.; Luque, F.J.; Lynch, C.I.; MacKerell, A.; Magistrato, A.; Marrink, S.J.; Martin, H.; McCammon, J.A.; Merz, K.; Moliner, V.; Mulholland, A.; Murad, S.; Naganathan, A.N.; Nangia, S.; Noe, F.; Noy, A.; Oláh, J.; O’Mara, M.; Ondrechen, M.J.; Onuchic, J.N.; Onufriev, A.; Osuna, S.; Panchenko, A.R.; Pantano, S.; Parish, C.; Parrinello, M.; Perez, A.; Perez-Acle, T.; Perilla, J.R.; Pettitt, B.M.; Pietropalo, A.; Piquemal, J.P.; Poma, A.; Praprotnik, M.; Ramos, M.J.; Ren, P.; Reuter, N.; Roitberg, A.; Rosta, E.; Rovira, C.; Roux, B.; Röthlisberger, U.; Sanbonmatsu, K.Y.; Schlick, T.; Shaytan, A.K.; Simmerling, C.; Smith, J.C.; Sugita, Y.; Świderek, K.; Taiji, M.; Tao, P.; Tikhonova, I.G.; Tirado-Rives, J.; Tuñón, I.; Kamp, M.W.V. d.; der Spoel, D.V.; Velankar, S.; Voth, G.A.; Wade, R.; Warshel, A.; Welborn, V.V.; Wetmore, S.; Wong, C.F.; Yang, L.W.; Zacharias, M.; Orozco, M.
The need to implement FAIR principles in biomolecular simulations.
Article page: https://www.nature.com/articles/s41592-025-02635-0
-
Chemistry - A European Journal, 2025, 31, e202403735.
Grajales-Hernández, D.A.; Roca, M.; Moliner, V.; López-Gallego, F.
-
ACS Synthetic Biology, 2025,
Bemelmans, M.; Wetzel, B.; Neusius, F.; Tieves, F.; Schwarz, C.; Mateljak, I.; Świderek, K.; Moliner, V.; Alcalde, M.; Sieber, V.
Article page: https://pubs.acs.org/doi/full/10.1021/acssynbio.5c00423
-
Chemical Science, 2025, 16, 14509.
Gabirondo, E.; Maiz-Iginitz, A.; Ximenis, M.; Świderek, K.; Andrés-Sanz, D.; Moliner, V.; Cabedo, L.; Westlie, A.H.; Chen, E.Y.X.; Cerrón-Infantes, A.; Unterlass, M.M.; Gallego, L.; Etxeberria, A.; Sardon, H.
Article page: https://pubs.rsc.org/en/content/articlelanding/2025/sc/d5sc02196k#!divAbstract
-
The Journal of Physical Chemistry Letters, 2025, 16, 3249.
Casalino, L.; Ramos-Guzmán, C.A.; Amaro, R.E.; Simmerling, C.; Lodola, A.; Mulholland, A.J.; Świderek, K.; Moliner, V.
A Reflection on the Use of Molecular Simulation to Respond to SARS-CoV-2 Pandemic Threats.
Article page: https://pubs.acs.org/doi/full/10.1021/acs.jpclett.4c03654
-
WIREs Computational Molecular Science , 2025, 15, e70003.
Świderek, K.; Bertran, J.; Zinovjev, K.; Tuñón, I.; Moliner, V.
Advances in the Simulations of Enzyme Reactivity in the Dawn of the Artificial Intelligence Age.
Article page: https://wires.onlinelibrary.wiley.com/doi/10.1002/wcms.70003
2024
-
Journal of Chemical Theory and Computation, 2024, 20, 1783.
Ruiz-Pernía, J.J.; Świderek, K.; Bertran, J.; Moliner, V.; Tuñón, I.
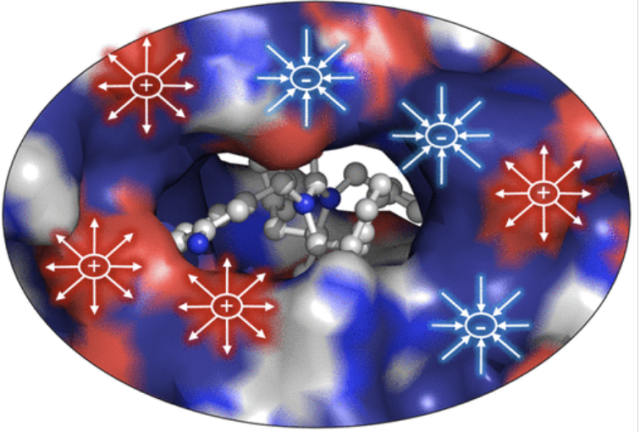
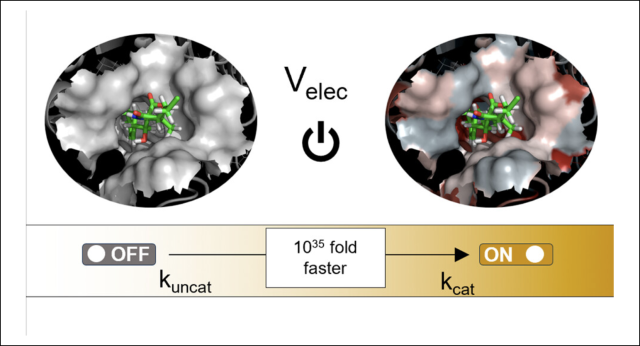
Electrostatics as a Guiding Principle in Understanding and Designing Enzymes.
Article page: https://pubs.acs.org/doi/full/10.1021/acs.jctc.3c01395
-
ACS Catalysis, 2024, 14, 15237.
Ferrer, S.; Moliner, V.; Świderek, K.
Electrostatic Preorganization in Three Distinct Heterogeneous Proteasome β-Subunits.
Article page: https://pubs.acs.org/doi/10.1021/acscatal.4c04964
-
Angewandte Chemie International Edition, 2024, 136, e202403098.
Williams, L.; Taily, I.M.; Hatton, L.; Berezin, A.A.; Wu, Y.L.; Moliner, V.; Świderek, K.; Tsai, Y.; Luk, L.Y.P.
Article page: https://onlinelibrary.wiley.com/doi/full/10.1002/ange.202403098
-
Faraday Discussions, 2024, 252, 323.
Świderek, K.; Martí, S.; Arafet, K.; Moliner, V.
Article page: https://pubs.rsc.org/en/content/articlehtml/2024/fd/d4fd00022f
-
Journal of Computational Chemistry, 2024, 20, 1783.
Ruiz-Pernía, J.J.; Świderek, K.; Bertran, J.; Moliner, V.; Tuñón, I.
Electrostatics as a Guiding Principle in Understanding and Designing Enzymes.
Article page: https://pubs.acs.org/doi/full/10.1021/acs.jctc.3c01395
-
Advance Science, 2024, 11, 2308956.
Gabirondo, E.; Świderek, K.; Marin, E.; Maiz-Iginitz, A.; Larranaga, A.; Moliner, V.; Etxeberria, A.; Sardon, H.
A Single Amino Acid Able to Promote High-Temperature Ring-Opening Polymerization by Dual Activation.
Article page: https://onlinelibrary.wiley.com/doi/10.1002/advs.202308956
-
Communications chemistry, 2024, 7, 15.
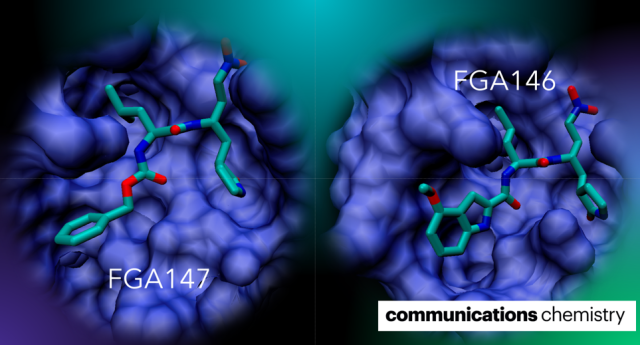
Medrano, F.J.; de la Hoz-Rodríguez, S.; Martí, S.; Arafet, K.; Schirmeister, T.; Hammerschmidt, S.J.; Müller, C.; González-Martínez, Á.; Santillana, E.; Ziebuhr, J.; Romero, A.; Zimmer, C.; Weldert, A.; Zimmermann, R.; Lodola, A.; Świderek, K.; Moliner, V.; González, F.V.
Article page: https://www.nature.com/articles/s42004-024-01104-7
2023
-
Journal of Chemical Informatics and Modeling, 2023, 63, 1301-1312.
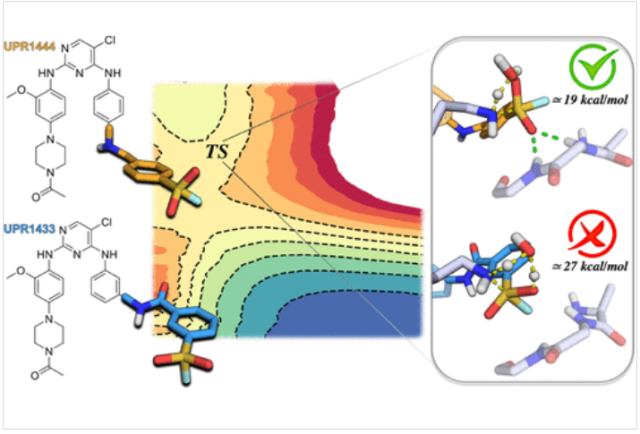
Arafet, K.; Scalvini, L.; Galvani, F.; Martí, S.; Moliner, V.; Mor, M.; Lodola, A.
-
Journal of American Chemical Society, 2023, 145,
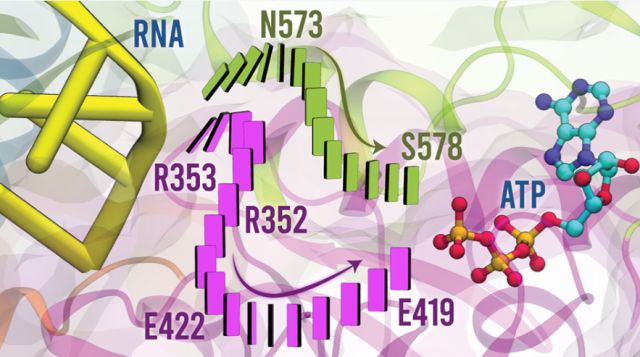
Movilla, S.; Roca, M.; Moliner, V.; Magistrato, A.
Molecular Basis of RNA-Driven ATP Hydrolysis in DExH-Box Helicases.
-
ACS Catalysis, 2023, 13, 6289-6300.
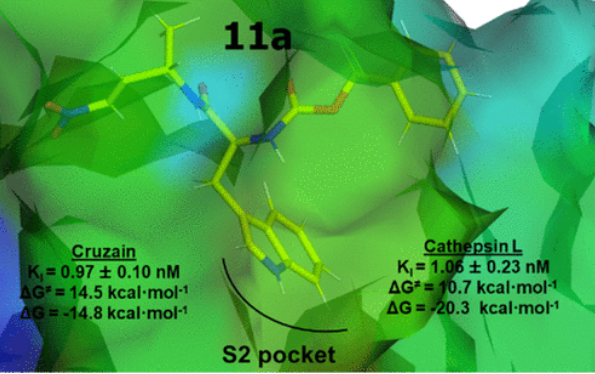
Arafet, K.; Royo, S.; Schirmeister, T.; Barthels, F.; González, F.V.; Moliner, V.
-
Nature Communications, 2023, 14, 3556.
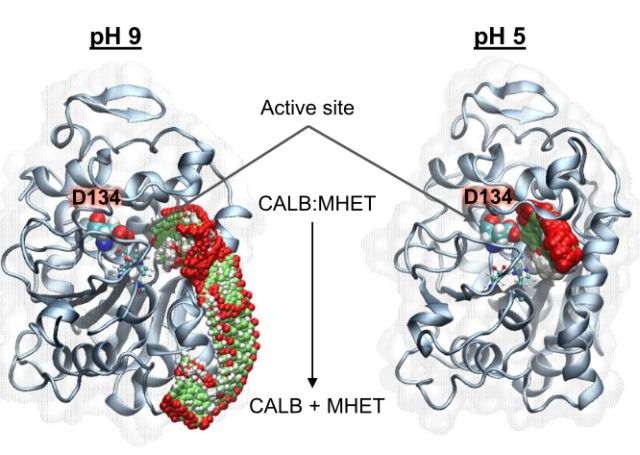
Świderek, K.; Velasco-Lozano, S.; Galmés, M.À.; Olazabal, I.; Sardon, H.; López-Gallego, F.; Moliner, V.
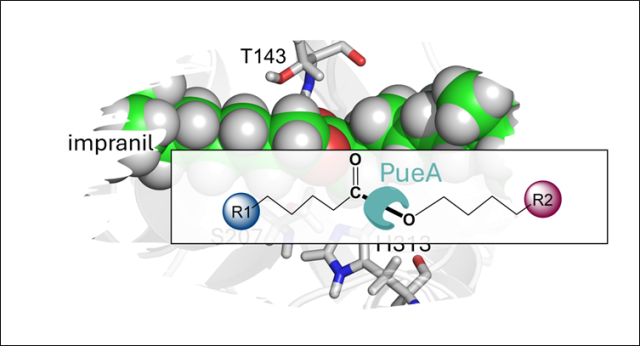
Mechanistic studies of a lipase unveil effect of pH on hydrolysis products of small PET modules.
-
ACS Catalysis, 2023, 13, 13354-13368.
Fernández-de-la-Pradilla, A.; Royo, S.; Schirmeister, T.; Barthels, F.; Świderek, K.; González, F.V.; Moliner, V.
-
Angewandte Chemie International Edition, 2023,
de Santos, G.; González-Benjumea, A.; Fernandez-Garcia, A.; Aranda, C.; Wu, Y.; But, A.; Molina-Espeja, P.; Maté, M.; Gonzalez-Perez, D.; Zhang, W.; Kiebist, J.; Scheibner, K.; Hofrichter, M.; Świderek, K.; Moliner, V.; Sanz-Aparicio, J.; Hollmann, F.; Gutiérrez, A.; Alcalde, M.
Engineering a Highly Regioselective Fungal Peroxygenase for the Synthesis of Hydroxy Fatty Acids.
-
Journal of Chemical Informatics and Modeling, 2023,
Movilla, S.; Martí, S.; Roca, M.; Moliner, V.