Research Overview
The BioComp group is devoted to the computational description of catalysis phenomena in biological systems, mostly enzyme catalyzed processes. For this purpose, theories, methods and techniques of theoretical and computational chemistry are developed, implemented and used. Computational algorithms and protocols, based on hybrid QM/MM potentials, are created and this new knowledge is exploited in designing new materials with properties to be used in biomedicine and biotechnology. In particular, these applications are focused on elucidating enzyme reaction mechanisms including fundamental new quantum and dynamical perspectives, to design new pharmacological agents to inhibit enzyme activity, or new catalysts mimicking the catalytic efficiency and features of natural enzymes.
Group's research interests
Development of QM/MM methods, combined with ML techniques, for the study of biochemical processes.
Deciphering the mode of action of enzymes.
Predicting enzymatic properties by computational simulations.(rate constants, Michaelis constants, inhibition constants, kinetic isotope effects, etc).
Design of new enzymes.
Design of new enzyme-inhibitors.
- Development of new biological materials with specific properties.
Members
Publications
2025
-
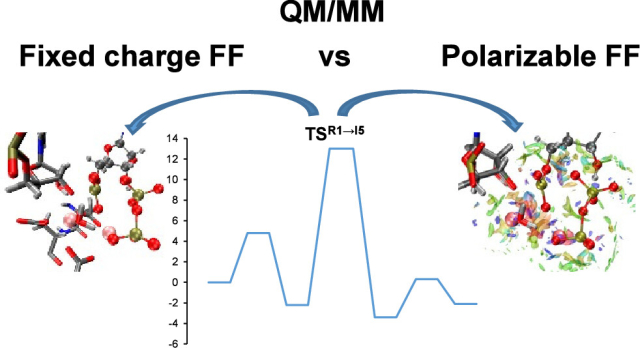
Journal of Chemical Theory and Computation, 2025, 21, 12342−12355.
Roca, M.; Maghsoud, Y.; Cisneros, G.A.; Świderek, K.; Moliner, V.
-
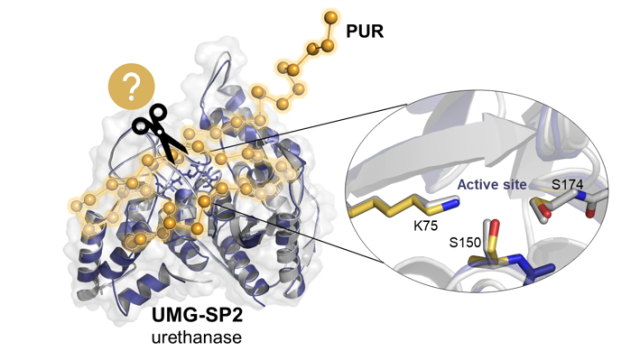
Journal of American Chemical Society, 2025, 147, 42511–42523.
Świderek, K.; Arafet, K.; Batista, Vde Sousa; Grajales-Hernández, D.; López-Gallego, F.; Moliner, V.
Insights into the Catalytic Activity of a Metagenome-Derived Urethanase.
Article page: https://pubs.acs.org/doi/10.1021/jacs.5c13147
-
Catalysis Science & Technology, 2025, 15, 6486-6498.
Pacheco, S.Fernando C.; Świderek, K.; Moran, M.Jesus; Oliden-Sánchez, A.; Moliner, V.; Salassa, L.; López-Gallego, F.
Overcoming Flavin-Driven Inactivation of Alcohol Dehydrogenases Through Enzyme Immobilization.
Article page: https://pubs.rsc.org/en/content/articlelanding/2025/cy/d5cy00964b
-
Nature Methods, 2025, 22, 641.
Amaro, R.E.; Åqvist, J.; Bahar, I.; Battistini, F.; Bellaiche, A.; Beltran, D.; Biggin, P.C.; Bonomi, M.; Bowman, G.R.; Bryce, R.; Bussi, G.; Carloni, P.; Case, D.; Cavalli, A.; Chang, C.E.A.; Cheung, M.S.; Chipot, C.; Chong, L.T.; Choudhary, P.; Cisneros, G.A.; Clementi, C.; Collepardo-Guevara, R.; Coveney, P.; Covino, R.; Crawford, T.D.; Peraro, M.D.; de Groot, B.; Delemotte, L.; Vivo, M.D.; Essex, J.; Fraternali, F.; Gao, J.; Gelpí, J.L.; Gervasio, F.L.; Gonzalez-Nilo, F.D.; Grubmüller, H.; Guenza, M.; Guzman, H.V.; Harris, S.; Head-Gordon, T.; Hernandez, R.; Hospital, A.; Huang, N.; Huang, X.; Hummer, G.; Iglesias-Fernández, J.; Jensen, J.H.; Jha, S.; Jiao, W.; Jorgensen, W.L.; Kamerlin, S.C.L.; Khalid, S.; Laughton, C.; Levitt, M.; Limongelli, V.; Lindahl, E.; Lindorff-Larsen, K.; Loverde, S.; Lundborg, M.; Luo, Y.L.; Luque, F.J.; Lynch, C.I.; MacKerell, A.; Magistrato, A.; Marrink, S.J.; Martin, H.; McCammon, J.A.; Merz, K.; Moliner, V.; Mulholland, A.; Murad, S.; Naganathan, A.N.; Nangia, S.; Noe, F.; Noy, A.; Oláh, J.; O’Mara, M.; Ondrechen, M.J.; Onuchic, J.N.; Onufriev, A.; Osuna, S.; Panchenko, A.R.; Pantano, S.; Parish, C.; Parrinello, M.; Perez, A.; Perez-Acle, T.; Perilla, J.R.; Pettitt, B.M.; Pietropalo, A.; Piquemal, J.P.; Poma, A.; Praprotnik, M.; Ramos, M.J.; Ren, P.; Reuter, N.; Roitberg, A.; Rosta, E.; Rovira, C.; Roux, B.; Röthlisberger, U.; Sanbonmatsu, K.Y.; Schlick, T.; Shaytan, A.K.; Simmerling, C.; Smith, J.C.; Sugita, Y.; Świderek, K.; Taiji, M.; Tao, P.; Tikhonova, I.G.; Tirado-Rives, J.; Tuñón, I.; Kamp, M.W.V. d.; der Spoel, D.V.; Velankar, S.; Voth, G.A.; Wade, R.; Warshel, A.; Welborn, V.V.; Wetmore, S.; Wong, C.F.; Yang, L.W.; Zacharias, M.; Orozco, M.
The need to implement FAIR principles in biomolecular simulations.
Article page: https://www.nature.com/articles/s41592-025-02635-0
-
Chemistry - A European Journal, 2025, 31, e202403735.
Grajales-Hernández, D.A.; Roca, M.; Moliner, V.; López-Gallego, F.
-
ACS Synthetic Biology, 2025, 14, 3721–3733.
Bemelmans, M.; Wetzel, B.; Neusius, F.; Tieves, F.; Schwarz, C.; Mateljak, I.; Świderek, K.; Moliner, V.; Alcalde, M.; Sieber, V.
Article page: https://pubs.acs.org/doi/full/10.1021/acssynbio.5c00423
-
Chemical Science, 2025, 16, 14509.
Gabirondo, E.; Maiz-Iginitz, A.; Ximenis, M.; Świderek, K.; Andrés-Sanz, D.; Moliner, V.; Cabedo, L.; Westlie, A.H.; Chen, E.Y.X.; Cerrón-Infantes, A.; Unterlass, M.M.; Gallego, L.; Etxeberria, A.; Sardon, H.
Article page: https://pubs.rsc.org/en/content/articlelanding/2025/sc/d5sc02196k#!divAbstract
-
The Journal of Physical Chemistry Letters, 2025, 16, 3249.
Casalino, L.; Ramos-Guzmán, C.A.; Amaro, R.E.; Simmerling, C.; Lodola, A.; Mulholland, A.J.; Świderek, K.; Moliner, V.
A Reflection on the Use of Molecular Simulation to Respond to SARS-CoV-2 Pandemic Threats.
Article page: https://pubs.acs.org/doi/full/10.1021/acs.jpclett.4c03654
-
WIREs Computational Molecular Science , 2025, 15, e70003.
Świderek, K.; Bertran, J.; Zinovjev, K.; Tuñón, I.; Moliner, V.
Advances in the Simulations of Enzyme Reactivity in the Dawn of the Artificial Intelligence Age.
Article page: https://wires.onlinelibrary.wiley.com/doi/10.1002/wcms.70003
2024
-
ACS Catalysis, 2024, 14, 15237.
Ferrer, S.; Moliner, V.; Świderek, K.
Electrostatic Preorganization in Three Distinct Heterogeneous Proteasome β-Subunits.
Article page: https://pubs.acs.org/doi/10.1021/acscatal.4c04964
-
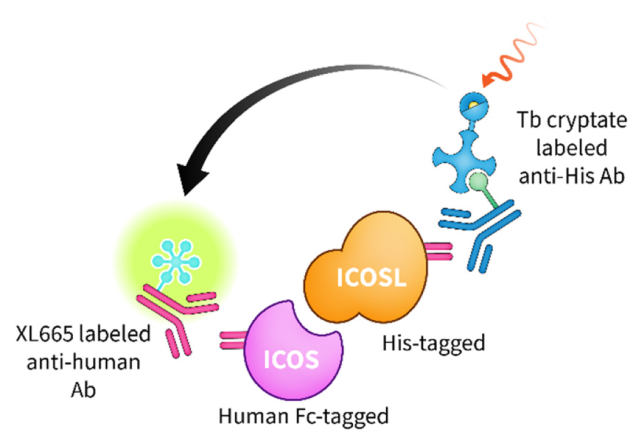
ChemMedChem, 2024, e202400545.
Zhang, L.; Calvo-Barreiro, L.; Batista, V.S.; Świderek, K.; Gabr, M.
Discovery of ICOS-targeted small molecules using affinity selection mass spectrometry screening.
Article page: https://chemistry-europe.onlinelibrary.wiley.com/doi/10.1002/cmdc.202400545
-
Angewandte Chemie International Edition, 2024, 136, e202403098.
Williams, L.; Taily, I.M.; Hatton, L.; Berezin, A.A.; Wu, Y.L.; Moliner, V.; Świderek, K.; Tsai, Y.; Luk, L.Y.P.
Article page: https://onlinelibrary.wiley.com/doi/full/10.1002/ange.202403098
-
Faraday Discussions, 2024, 252, 323.
Świderek, K.; Martí, S.; Arafet, K.; Moliner, V.
Article page: https://pubs.rsc.org/en/content/articlehtml/2024/fd/d4fd00022f
-
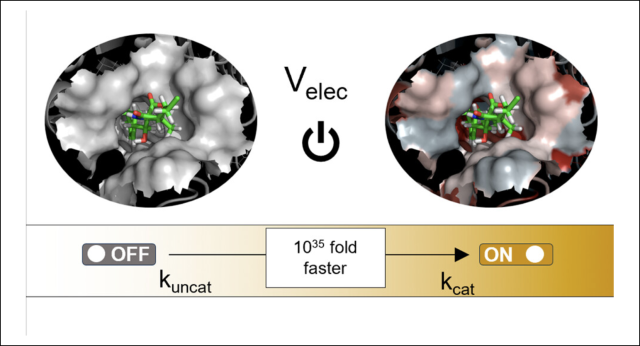
Journal of Computational Chemistry, 2024, 20, 1783.
Ruiz-Pernía, J.J.; Świderek, K.; Bertran, J.; Moliner, V.; Tuñón, I.
Electrostatics as a Guiding Principle in Understanding and Designing Enzymes.
Article page: https://pubs.acs.org/doi/full/10.1021/acs.jctc.3c01395
-
Advance Science, 2024, 11, 2308956.
Gabirondo, E.; Świderek, K.; Marin, E.; Maiz-Iginitz, A.; Larranaga, A.; Moliner, V.; Etxeberria, A.; Sardon, H.
A Single Amino Acid Able to Promote High-Temperature Ring-Opening Polymerization by Dual Activation.
Article page: https://onlinelibrary.wiley.com/doi/10.1002/advs.202308956
-
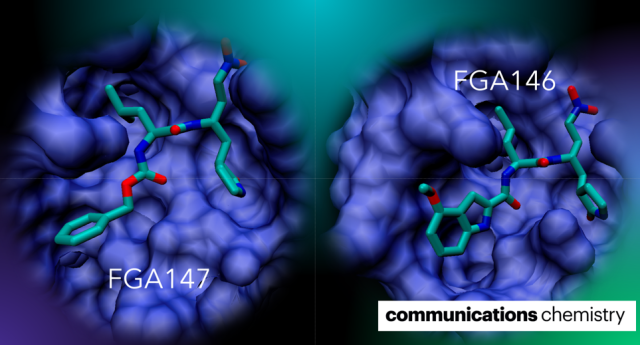
Communications chemistry, 2024, 7, 15.
Medrano, F.J.; de la Hoz-Rodríguez, S.; Martí, S.; Arafet, K.; Schirmeister, T.; Hammerschmidt, S.J.; Müller, C.; González-Martínez, Á.; Santillana, E.; Ziebuhr, J.; Romero, A.; Zimmer, C.; Weldert, A.; Zimmermann, R.; Lodola, A.; Świderek, K.; Moliner, V.; González, F.V.
Article page: https://www.nature.com/articles/s42004-024-01104-7
2023
-
RSC Medicinal Chemistry, 2023, 14, 1767-1777.
Abdel-Rahman, S.A.; Świderek, K.; Gabr, M.T.
Article page: https://pubs.rsc.org/en/content/articlelanding/2023/md/d3md00150d
-
Journal of Chemical Informatics and Modeling, 2023, 63, 1301-1312.
Arafet, K.; Scalvini, L.; Galvani, F.; Martí, S.; Moliner, V.; Mor, M.; Lodola, A.
-
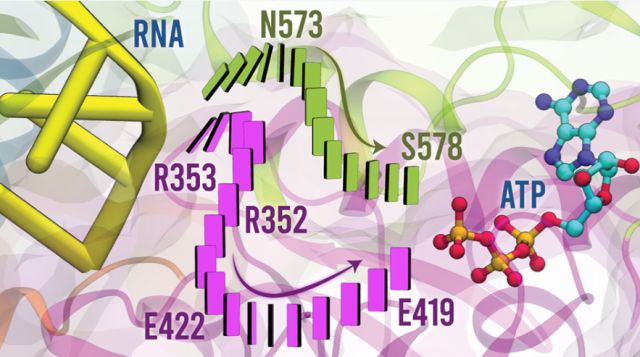
Journal of American Chemical Society, 2023, 145,
Movilla, S.; Roca, M.; Moliner, V.; Magistrato, A.
Molecular Basis of RNA-Driven ATP Hydrolysis in DExH-Box Helicases.
-
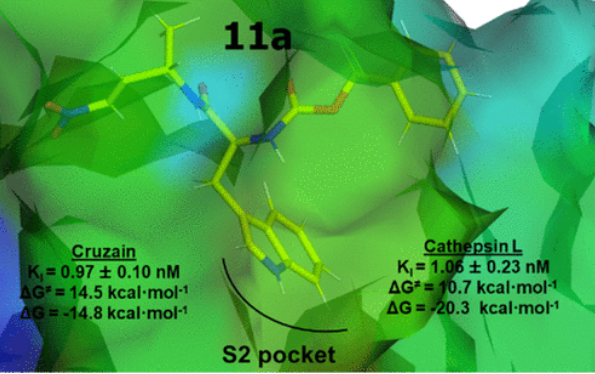
ACS Catalysis, 2023, 13, 6289-6300.
Arafet, K.; Royo, S.; Schirmeister, T.; Barthels, F.; González, F.V.; Moliner, V.